Plant synthetic biology as a tool to understand and engineer

Since my beginnings I’ve been a stubborn and relentless plant enthusiast. Born in the farmlands of Pennsylvania, I was steeped in the local ecology and how agriculture integrates into the landscape. Once I found out that it would require a firm foundation in molecular biology to make Jurassic park-like scenarios a reality, I decided research was the path for me.
I followed my love for plant biology to research in plant reproduction in Graduate school at Brown University in the Lab of Dr. Mark Johnson. We discovered how a single-celled pollen grain undergoes differentiation during reproduction to deliver the sperm to the egg, through activation of a subset of transcription factors (see this movie for more information).
From this work I came to appreciate the combinatory interaction network of transcriptional regulators that must perfectly coordinate to create molecular identity and designate the ‘self’ identify. How do cells interpret their who and where they are in a cellular environment? How does a cell compute the mixture of signals into a behavior - and how does that behavior contribute to a whole organism?
Pollen grains are tasked with running a gauntlet that determines their fitness to deliver sperm to the egg. In vivo, they grow through the stigma, style and into the ovules delivering the sperm (red cells) to the egg cells buried inside the ovule. Here, I have exposed the scene by dissecting the stigma/style (top middle) onto media to allow pollen tubes to run over a flat agar plate to dissected ovules. The expression of a group of pollen tube specific transcription factors (here visualized in the green channel, MYB97:GFP) is critical to differentiate the pollen tube to be ready for interaction with the ovule.

Fascinated by the molecular control over cellular differentiation I observed in my graduate studies, I decided to study how networks are engineered. Therefore, I went to the laboratory of Dr. Jennifer Nemhauser at the University of Washington. Synthetic biology allows us to decode molecular process by engineering them, as by attempting to engineer them from scratch identifies our gaps in knowledge of the system. Bringing synthetic biology approaches to plants presents pathways to mitigating the worst effects of climate change, and to engineer crops for destroyed environments on earth or entirely novel environments such as those encountered in space travel.
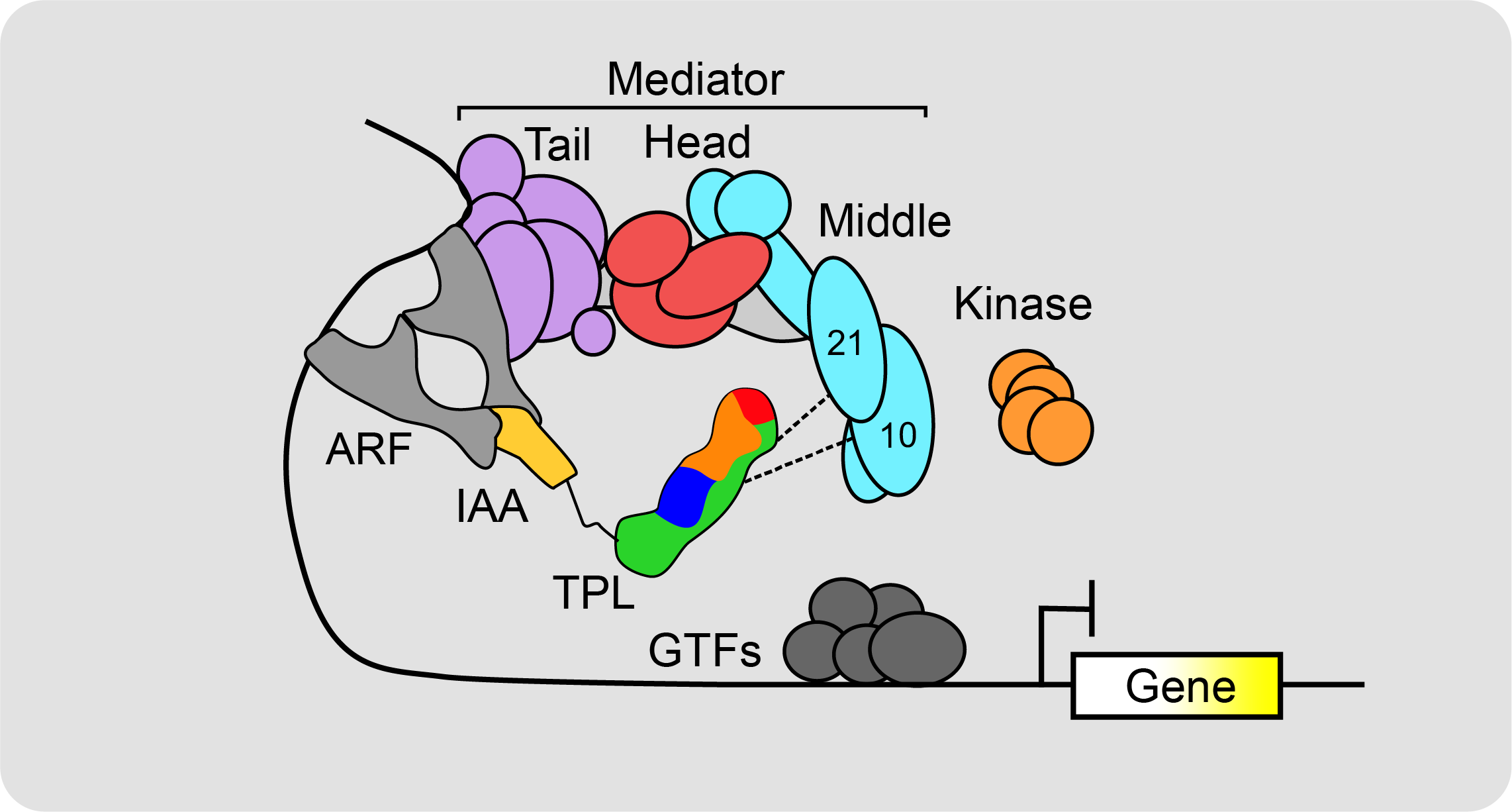
The ability to engineer new traits using synthetic signaling methods requires standardized libraries of biological parts and rapid methods to assemble them; the decoupling of complex processes into simpler subsystems; and mathematical models that can accelerate the design-build-test-learn cycle. In the Nemhauser Lab, I have been working towards expanding our understanding and repertoire of transcriptional repressors to effectively tune eukaryotic transcriptional output. Focusing on the conserved Groucho/Tup1-type family in plants known as TOPLESS (TPL), I have leveraged structural knowledge to identify the protein domains responsible for repression, and defined one modality of TPL’s transcriptional repression acts by inhibiting Mediator complex activity.
Publications
Repression by the Arabidopsis TOPLESS corepressor requires association with the core mediator complex. Leydon AR, Wang W, Gala HP, Gilmour S, Juarez-Solis S, Zahler ML, Zemke JE, Zheng N, Nemhauser JL. Elife. 2021 June 2;10:e66739. doi:10.7554/eLife.66739. PMID: 34075876; PMCID: PMC8203292.
Engineering Synthetic Signaling in Plants. Leydon AR, Gala HP, Guiziou S, Nemhauser JL. Annu Rev Plant Biol. 2020 Apr 29;71:767-788. doi: 10.1146/annurev-arplant-081519-035852. Epub 2020 Feb 24. PMID: 32092279.
Accelerating structure-function mapping using the ViVa webtool to mine natural variation. Hamm MO, Moss BL, Leydon AR, Gala HP, Lanctot A, Ramos R, Klaeser H, Lemmex AC, Zahler ML, Nemhauser JL, Wright RC. Plant Direct. 2019 Jul 26;3(7):e00147. doi: 10.1002/pld3.147. PMID: 31372596; PMCID: PMC6658840.
Synthetic hormone-responsive transcription factors can monitor and re-program plant development. Khakhar A, Leydon AR, Lemmex AC, Klavins E, Nemhauser JL. Elife. 2018 May 1;7:e34702. doi: 10.7554/eLife.34702. PMID: 29714687; PMCID: PMC5976440.
The Molecular Dialog between Flowering Plant Reproductive Partners Defined by SNP-Informed RNA-Sequencing. Leydon AR, Weinreb C, Venable E, Reinders A, Ward JM, Johnson MA. Plant Cell. 2017 May;29(5):984-1006. doi: 10.1105/tpc.16.00816. Epub 2017 Apr 11. PMID: 28400492; PMCID: PMC5466024.
Transporters involved in pH and K+ homeostasis affect pollen wall formation, male fertility, and embryo development. Padmanaban S, Czerny DD, Levin KA, Leydon AR, Su RT, Maugel TK, Zou Y, Chanroj S, Cheung AY, Johnson MA, Sze H. J Exp Bot. 2017 Jun 1;68(12):3165-3178. doi: 10.1093/jxb/erw483. PMID: 28338823; PMCID: PMC5853877.
Pollen Tube Discharge Completes the Process of Synergid Degeneration That Is Initiated by Pollen Tube-Synergid Interaction in Arabidopsis. Leydon AR, Tsukamoto T, Dunatunga D, Qin Y, Johnson MA, Palanivelu R. Plant Physiol. 2015 Sep;169(1):485-96. doi: 10.1104/pp.15.00528. Epub 2015 Jul 30. PMID: 26229050; PMCID: PMC4577395.
Interactions between pollen tube and pistil control pollen tube identity and sperm release in the Arabidopsis female gametophyte. Leydon AR, Chaibang A, Johnson MA. Biochem Soc Trans. 2014 Apr;42(2):340-5. doi: 10.1042/BST20130223. PMID: 24646241; PMCID: PMC5512271.
The CLAMP protein links the MSL complex to the X chromosome during Drosophila dosage compensation. Soruco MM, Chery J, Bishop EP, Siggers T, Tolstorukov MY, Leydon AR, Sugden AU, Goebel K, Feng J, Xia P, Vedenko A, Bulyk ML, Park PJ, Larschan E. Genes Dev. 2013 Jul 15;27(14):1551-6. doi: 10.1101/gad.214585.113. PMID: 23873939; PMCID: PMC3731544.
Three MYB transcription factors control pollen tube differentiation required for sperm release. Leydon AR, Beale KM, Woroniecka K, Castner E, Chen J, Horgan C, Palanivelu R, Johnson MA. Curr Biol. 2013 Jul 8;23(13):1209-14. doi: 10.1016/j.cub.2013.05.021. Epub 2013 Jun 20. PMID: 23791732; PMCID: PMC4009620.
Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Beale KM, Leydon AR, Johnson MA. Curr Biol. 2012 Jun 19;22(12):1090-4. doi: 10.1016/j.cub.2012.04.041. Epub 2012 May 17. PMID: 22608506; PMCID: PMC3973743.
HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. Wong JL, Leydon AR, Johnson MA. PLoS Genet. 2010 Mar 19;6(3):e1000882. doi: 10.1371/journal.pgen.1000882. PMID: 20333238; PMCID: PMC2841615.
Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R. PLoS Genet. 2009 Aug;5(8):e1000621. doi: 10.1371/journal.pgen.1000621. Epub 2009 Aug 28. Erratum in: PLoS Genet. 2016 Jul;12(7):e1006210. PMID: 19714218; PMCID: PMC2726614.
Late nights in lab.
Link to blog posts maybe?

Contact me.
aleydon at uw.edu
shipping address:
department of biology, university of washington, box 351800, seattle, wa 98195-1800